An R Script to Automatically download PubMed Citation Counts By Year of publication
Background
I believe there’s some information to be gained from looking at publication trends over time. But it’s really troublesome to do it by hand; fortunately it’s not so troublesome to do it in R statistical software. Though, data like these should be interpreted with extreme caution.
How the script works
I tried to use the RISmed-package to query PubMed, but found it to be
really unreliable. Instead my script is querying PubMed’s
E-Utilities using RCurl and XML. The E-utilities work like this:
http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&rettype=count&term=search+termWe can see after the URL that I’m telling E-Utilities that I want to search PubMed’s database, and retrieve it as ‘count’. This will give a minimal XML-output containing only the number of hits for my search term, which is exactly what I want.
<eSearchResult><Count>140641</Count></eSearchResult>That’s really the basic gist of what my script is doing. If you look at
the code (at the bottom of this post), you can see that I construct the
query in the beginning of the script using paste() and gsub(). Then
the main part of the script is the for-loop. What it’s doing is it’s
looping through all the years from the specified year range, retrieving
the number of hits, and then pasting it together in one data frame. To
get counts for a specific year i, I add AND i[PPDAT] (Print Dates
Only Tag) at the end of each query.
Since I have all the necessary code in getCount(), I can run the same
script for any number of queries using ldply(query.index, getCount).
By doing that I end up with a data frame, which contains data for all
the queries arranged in long format. The end of the script will
calculate relative counts by dividing the matches each year by the
total amount of publications that year. I’ve also added a function
that will type out the total number of hits for each query, called
PubTotalHits().
Why use Print Dates Only Tag [PPDAT] and not Publication Date [DP]?
The problem with using DP is in how PubMed handles articles. If an article is published electronically in the end of say 2011 but printed in 2012, that article will be counted both in 2011 and 2012 if I search those 2 years individually (which my script is doing).
By using PPDAT I will miss some articles that doesn’t have a published print date. If you’d rather get some duplicates in your data, but not miss any citations, you can easily change PPDAT to DP, the script will run the same either way.
A quick example [PDAT] vs [DP]:
To illustrate the differences I did a quick search using a Cognitive Behavioral Therapy-query. When searching with PubMed’s website I specified the year range as 1940:2012[DP]/[PPDAT], and used the same interval in R.
PPDAT: Pubmed 5372 R 5372 DP: Pubmed 5501 R 5661The correct amount of hits is 5501 which is retrieved using PubMed’s website with the [DP]-tag. It’s also the same amount as what would be reported when not specifying any time interval. Consequently, if you use my script with the [PPDAT]-tag you would, in this scenario, be about -2.5% off from the correct amount, and about +3% off from the correct amount if you use [DP]. It’s possible that other queries will generate different results. However, the error seems to be so small that it doesn’t warrant any changes to the code. Duplicates could be avoided by downloading PMIDS with every search, then checking for duplicates for each adjacent year. Though, that change would require an unnecessary amount of data transfer for an error that appear to be only 3%.
How to use my script
It’s really simple to use this script. Assuming you have R installed, all you need to do is download the files. Then point R to that directory and then tell it to run “PubMedTrend.R”. Like this:
setwd("/path/to/directory")# Script sourcesource("PubMedTrend.R")Once that is done you specify your query like this:
query <- c("name of query" = "actually query", "ssri" = "selective serotonin reuptake inhibitor*[tiab]")Now all you have to is to execute my PubMedTrend()-function for those
quires and save the results in a data frame:
df <- PubMedTrend(query)The content of df will be structured like this:
> head(df) .id year count total_count relative1 cbt 1970 1 212744 0.047004852 cbt 1976 2 249328 0.080215623 cbt 1977 1 255863 0.039083424 cbt 1978 1 265591 0.037651885 cbt 1979 1 274302 0.036456176 cbt 1980 9 272549 0.33021585Additional arguments
The default year range is set to 1950–2009, but can easily be changed, like this:
df <- PubMedTrend(query, 1990,2005)Some notes about using relative values
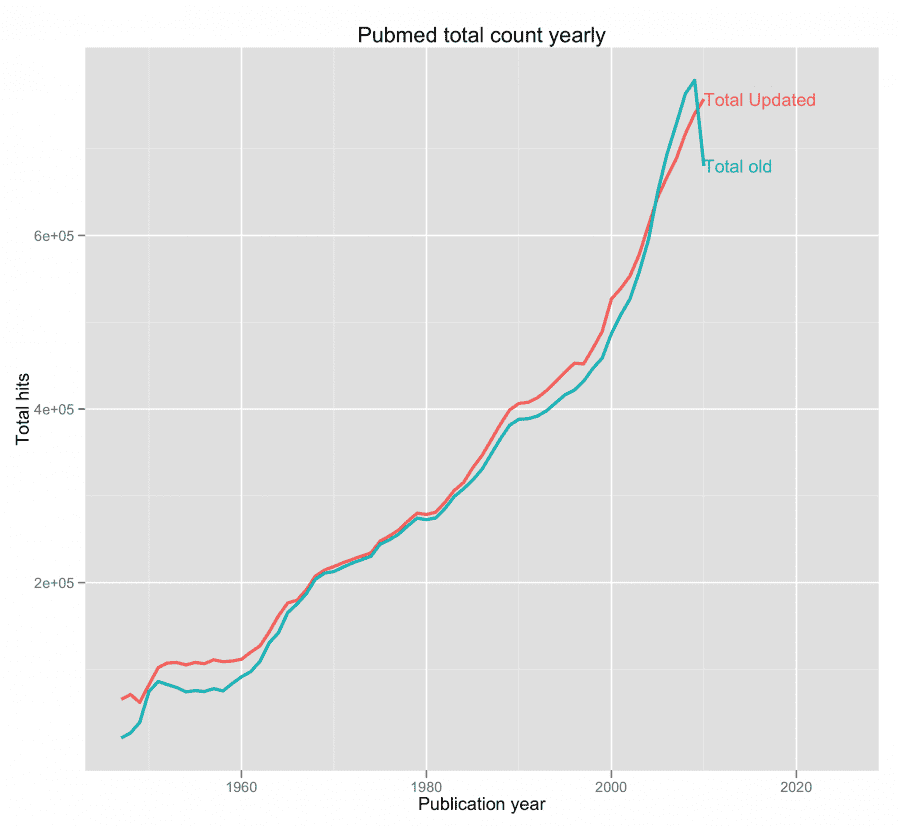
PubMed’s total counts (as posted in a table at their website) hasn’t been updated since April 8 2011, but the de facto total values have changed since then, because PubMed is always adding new citations (new and old). This can be remedied easily by looping through 1950[PPDAT], 1951[PPDAT] … 2012[PPDAT] (or you can use [DP]). I did that for you and made a graph of the two data sets, and as you can see there’s some differences, but they’re not that big. Nonetheless, I’ve included both files with my script.
Some example runs
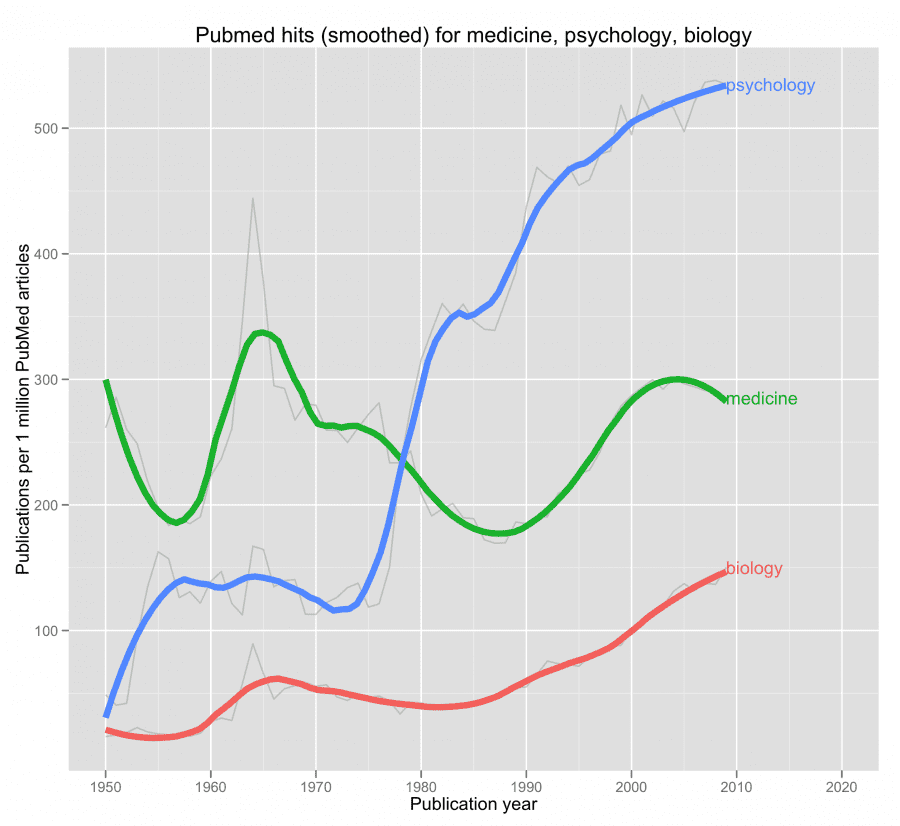
query <- c("medicine" = "medicine[tw]","psychology" = "psychology[tw]","biology" = "biology[tw]")
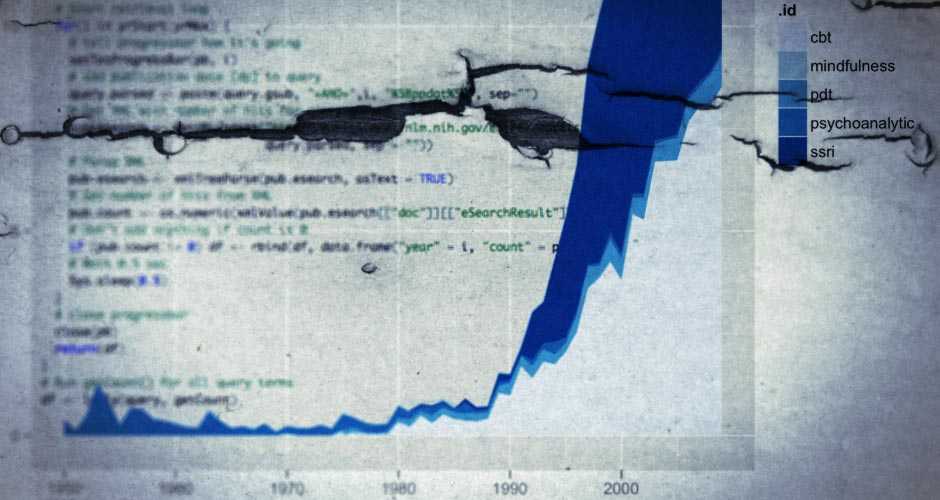
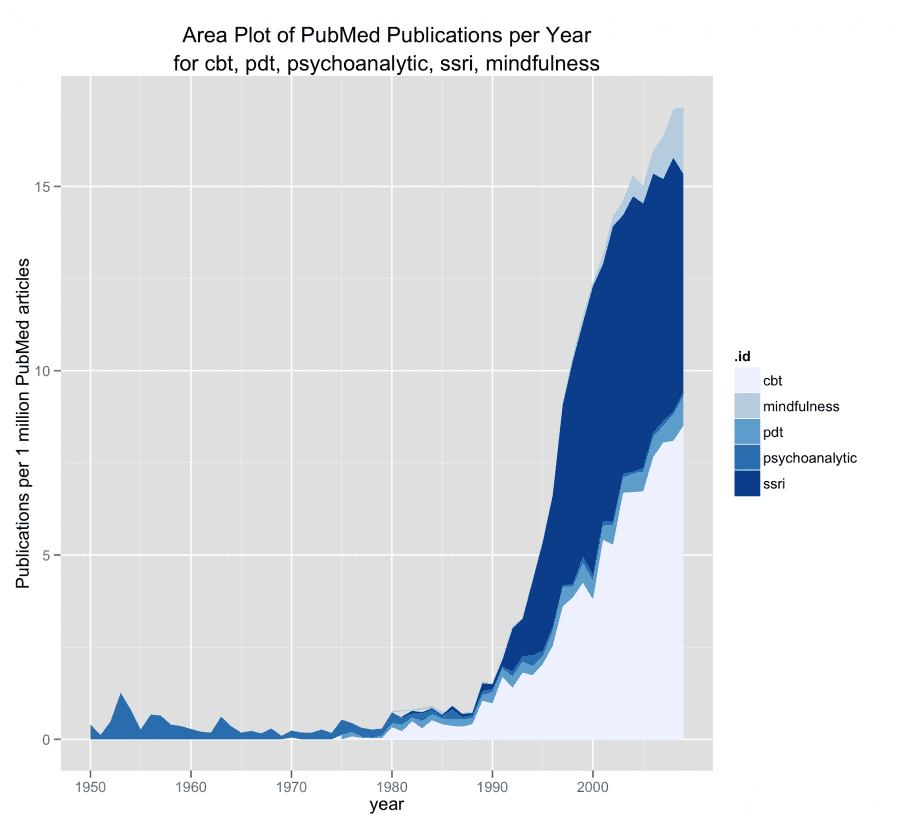
query <- c("cbt"= "cognitive behav* psychotherap*[tiab] OR cognitive behav* therap*[tiab]", "pdt" = "psychodynamic therap*[tiab] OR psychodynamic psychotherap*[tiab]", "psychoanalytic" = "psychoanalytic therap*[tiab] OR psychoanalytic psychoterap*[tiab]", "ssri" = "selective serotonin reuptake inhibitor*[tiab]", "mindfulness" = "mindfulness[tiab]")
When searching a progress bar will show the progress, and the search will look like this once completed.
> df <- PubMedTrend(query)Searching for: cognitive behav* psychotherap*[tiab] OR cognitive behav* therap*[tiab] |=======================================================================================================================| 100%Searching for: psychodynamic therap*[tiab] OR psychodynamic psychotherap*[tiab] |=======================================================================================================================| 100%Searching for: psychoanalytic therap*[tiab] OR psychoanalytic psychoterap*[tiab] |=======================================================================================================================| 100%Searching for: psychotherapy[tiab] |=======================================================================================================================| 100%Searching for: selective serotonin reuptake inhibitor*[tiab] |=======================================================================================================================| 100%Searching for: mindfulness[tiab] |=======================================================================================================================| 100%All done!Using the function to get total hits, will give this output
> PubTotalHits() search_name query total_hits1 cbt cognitive behav* psychotherap*[tiab] OR cognitive behav* therap*[tiab] 55142 pdt psychodynamic therap*[tiab] OR psychodynamic psychotherap*[tiab] 5543 psychoanalytic psychoanalytic therap*[tiab] OR psychoanalytic psychoterap*[tiab] 2944 psychotherapy psychotherapy[tiab] 208435 ssri selective serotonin reuptake inhibitor*[tiab] 56536 mindfulness mindfulness[tiab] 529A few words on usage guidelines
In PubMed’s E-utilities usage guidelines it’s specified that:
In order not to overload the E-utility servers, NCBI recommends that users post no more than three URL requests per second and limit large jobs to either weekends or between 9:00 PM and 5:00 AM Eastern time during weekdays. Failure to comply with this policy may result in an IP address being blocked from accessing NCBI
To comply with this my script will wait 0.5 sec after each iteration resulting in (theoretically) 2 URL GETs per second. This means that searching for 100 yearly counts will take a minimum if 50 seconds for each query. You can change the wait time if you feel that 0.5 sec is too low or too high.
And here’s the R code to look at PubMed trends
Update (2013 August 2): I am currently updating this script and moving it to a GitHub repo, so it will be easier to maintain. You can find the repo here.
And some example ggplot2 codes
# Note:# ——# These plots wont work if you only have 1 query. ### AREA PLOT ###ggplot(df, aes(year, relative, group=.id, fill=.id)) +geom_area() +opts(title=paste("Area Plot of PubMed Publications per Year\nfor", paste(names(query), collapse = ", "))) +xlab("year") +ylab("Publications per 1 million PubMed articles") +scale_fill_brewer() ### LINE PLOTS ### # RAWggplot(df, aes(year, relative, group=.id, color=.id)) +geom_line(show_guide=F) +xlab("Publication year") +ylab("Publications per 1 million PubMed articles") +opts(title = paste("Pubmed hits for", paste(names(query), collapse = ", "))) # SMOOTHEDp <- ggplot(df, aes(year, relative, group=.id, color=.id)) +geom_line(alpha = I(7/10), color="grey", show_guide=F) +stat_smooth(size=2, span=0.3, se=F, show_guide=F) +xlab("Publication year") +ylab("Publications per 1 million PubMed articles") +opts(title = paste("Pubmed hits (smoothed) for", paste(names(query), collapse = ", "))) +xlim(1950,2020)#direct.label(p, "last.bumpup")Written by Kristoffer Magnusson, a researcher in clinical psychology. You should follow him on Bluesky or on Twitter.
Published April 19, 2012 (View on GitHub)
Buy Me A Coffee
A huge thanks to the 175 supporters who've bought me a 422 coffees!
Steffen bought ☕☕☕☕☕☕☕☕☕☕☕☕ (12) coffees
I love your visualizations. Some of the best out there!!!
Jason Rinaldo bought ☕☕☕☕☕☕☕☕☕☕ (10) coffees
I've been looking for applets that show this for YEARS, for demonstrations for classes. Thank you so much! Students do not need to tolarate my whiteboard scrawl now. I'm sure they'd appreciate you, too.l
Shawn Bergman bought ☕☕☕☕☕ (5) coffees
Thank you for putting this together! I am using these visuals and this information to teach my Advanced Quant class.
anthonystevendick@gmail.com bought ☕☕☕☕☕ (5) coffees
I've been using a lot of your ideas in a paper I'm writing and even borrowed some of your code (cited of course). But this site has been so helpful I think, in addition, I owe you a few coffees!
Chip Reichardt bought ☕☕☕☕☕ (5) coffees
Hi Krisoffer, these are great applets and I've examined many. I'm writing a chapter for the second edition of "Teaching statistics and quantitative methods in the 21st century" by Joe Rodgers (Routledge). My chapter is on the use of applets in teaching statistics. I could well be describing 5 of yours. Would you permit me to publish one or more screen shots of the output from one or more of your applets. I promise I will be saying very positive things about your applets. If you are inclined to respond, my email address if Chip.Reichardt@du.edu.
Someone bought ☕☕☕☕☕ (5) coffees
Someone bought ☕☕☕☕☕ (5) coffees
Nice work! Saw some of your other publications and they are also really intriguing. Thanks so much!
JDMM bought ☕☕☕☕☕ (5) coffees
You finally helped me understand correlation! Many, many thanks... 😄
@VicCazares bought ☕☕☕☕☕ (5) coffees
Good stuff! It's been so helpful for teaching a Psych Stats class. Cheers!
Dustin M. Burt bought ☕☕☕☕☕ (5) coffees
Excellent and informative visualizations!
Someone bought ☕☕☕☕☕ (5) coffees
@metzpsych bought ☕☕☕☕☕ (5) coffees
Always the clearest, loveliest simulations for complex concepts. Amazing resource for teaching intro stats!
Ryo bought ☕☕☕☕☕ (5) coffees
For a couple years now I've been wanting to create visualizations like these as a way to commit these foundational concepts to memory. But after finding your website I'm both relieved that I don't have to do that now and pissed off that I couldn't create anything half as beautiful and informative as you have done here. Wonderful job.
Diarmuid Harvey bought ☕☕☕☕☕ (5) coffees
You have an extremely useful site with very accessible content that I have been using to introduce colleagues and students to some of the core concepts of statistics. Keep up the good work, and thanks!
Michael Hansen bought ☕☕☕☕☕ (5) coffees
Keep up the good work!
Michael Villanueva bought ☕☕☕☕☕ (5) coffees
I wish I could learn more from you about stats and math -- you use language in places that I do not understand. Cohen's D visualizations opened my understanding. Thank you
Someone bought ☕☕☕☕☕ (5) coffees
Thank you, Kristoffer
Pål from Norway bought ☕☕☕☕☕ (5) coffees
Great webpage, I use it to illustrate several issues when I have a lecture in research methods. Thanks, it is really helpful for the students:)
@MAgrochao bought ☕☕☕☕☕ (5) coffees
Joseph Bulbulia bought ☕☕☕☕☕ (5) coffees
Hard to overstate the importance of this work Kristoffer. Grateful for all you are doing.
@TDmyersMT bought ☕☕☕☕☕ (5) coffees
Some really useful simulations, great teaching resources.
@lakens bought ☕☕☕☕☕ (5) coffees
Thanks for fixing the bug yesterday!
@LinneaGandhi bought ☕☕☕☕☕ (5) coffees
This is awesome! Thank you for creating these. Definitely using for my students, and me! :-)
@ICH8412 bought ☕☕☕☕☕ (5) coffees
very useful for my students I guess
@KelvinEJones bought ☕☕☕☕☕ (5) coffees
Preparing my Master's student for final oral exam and stumbled on your site. We are discussing in lab meeting today. Coffee for everyone.
Someone bought ☕☕☕☕☕ (5) coffees
What a great site
@Daniel_Brad4d bought ☕☕☕☕☕ (5) coffees
Wonderful work!
David Loschelder bought ☕☕☕☕☕ (5) coffees
Terrific work. So very helpful. Thank you very much.
@neilmeigh bought ☕☕☕☕☕ (5) coffees
I am so grateful for your page and can't thank you enough!
@giladfeldman bought ☕☕☕☕☕ (5) coffees
Wonderful work, I use it every semester and it really helps the students (and me) understand things better. Keep going strong.
Dean Norris bought ☕☕☕☕☕ (5) coffees
Sal bought ☕☕☕☕☕ (5) coffees
Really super useful, especially for teaching. Thanks for this!
dde@paxis.org bought ☕☕☕☕☕ (5) coffees
Very helpful to helping teach teachers about the effects of the Good Behavior Game
@akreutzer82 bought ☕☕☕☕☕ (5) coffees
Amazing visualizations! Thank you!
@rdh_CLE bought ☕☕☕☕☕ (5) coffees
So good!
tchipman1@gsu.edu bought ☕☕☕ (3) coffees
Hey, your stuff is cool - thanks for the visual
Hugo Quené bought ☕☕☕ (3) coffees
Hi Kristoffer, Some time ago I've come up with a similar illustration about CIs as you have produced, and I'm now also referring to your work:<br>https://hugoquene.github.io/QMS-EN/ch-testing.html#sec:t-confidenceinterval-mean<br>With kind regards, Hugo Quené<br>(Utrecht University, Netherlands)
Tor bought ☕☕☕ (3) coffees
Thanks so much for helping me understand these methods!
Amanda Sharples bought ☕☕☕ (3) coffees
Soyol bought ☕☕☕ (3) coffees
Someone bought ☕☕☕ (3) coffees
Kenneth Nilsson bought ☕☕☕ (3) coffees
Keep up the splendid work!
@jeremywilmer bought ☕☕☕ (3) coffees
Love this website; use it all the time in my teaching and research.
Someone bought ☕☕☕ (3) coffees
Powerlmm was really helpful, and I appreciate your time in putting such an amazing resource together!
DR AMANDA C DE C WILLIAMS bought ☕☕☕ (3) coffees
This is very helpful, for my work and for teaching and supervising
Georgios Halkias bought ☕☕☕ (3) coffees
Regina bought ☕☕☕ (3) coffees
Love your visualizations!
Susan Evans bought ☕☕☕ (3) coffees
Thanks. I really love the simplicity of your sliders. Thanks!!
@MichaMarie8 bought ☕☕☕ (3) coffees
Thanks for making this Interpreting Correlations: Interactive Visualizations site - it's definitely a great help for this psych student! 😃
Zakaria Giunashvili, from Georgia bought ☕☕☕ (3) coffees
brilliant simulations that can be effectively used in training
Someone bought ☕☕☕ (3) coffees
@PhysioSven bought ☕☕☕ (3) coffees
Amazing illustrations, there is not enough coffee in the world for enthusiasts like you! Thanks!
Cheryl@CurtinUniAus bought ☕☕☕ (3) coffees
🌟What a great contribution - thanks Kristoffer!
vanessa moran bought ☕☕☕ (3) coffees
Wow - your website is fantastic, thank you for making it.
Someone bought ☕☕☕ (3) coffees
mikhail.saltychev@gmail.com bought ☕☕☕ (3) coffees
Thank you Kristoffer This is a nice site, which I have been used for a while. Best Prof. Mikhail Saltychev (Turku University, Finland)
Someone bought ☕☕☕ (3) coffees
Ruslan Klymentiev bought ☕☕☕ (3) coffees
@lkizbok bought ☕☕☕ (3) coffees
Keep up the nice work, thank you!
@TELLlab bought ☕☕☕ (3) coffees
Thanks - this will help me to teach tomorrow!
SCCT/Psychology bought ☕☕☕ (3) coffees
Keep the visualizations coming!
@elena_bolt bought ☕☕☕ (3) coffees
Thank you so much for your work, Kristoffer. I use your visualizations to explain concepts to my tutoring students and they are a huge help.
A random user bought ☕☕☕ (3) coffees
Thank you for making such useful and pretty tools. It not only helped me understand more about power, effect size, etc, but also made my quanti-method class more engaging and interesting. Thank you and wish you a great 2021!
@hertzpodcast bought ☕☕☕ (3) coffees
We've mentioned your work a few times on our podcast and we recently sent a poster to a listener as prize so we wanted to buy you a few coffees. Thanks for the great work that you do!Dan Quintana and James Heathers - Co-hosts of Everything Hertz
Cameron Proctor bought ☕☕☕ (3) coffees
Used your vizualization in class today. Thanks!
eshulman@brocku.ca bought ☕☕☕ (3) coffees
My students love these visualizations and so do I! Thanks for helping me make stats more intuitive.
Someone bought ☕☕☕ (3) coffees
Adrian Helgå Vestøl bought ☕☕☕ (3) coffees
@misteryosupjoo bought ☕☕☕ (3) coffees
For a high school teacher of psychology, I would be lost without your visualizations. The ability to interact and manipulate allows students to get it in a very sticky manner. Thank you!!!
Chi bought ☕☕☕ (3) coffees
You Cohen's d post really helped me explaining the interpretation to people who don't know stats! Thank you!
Someone bought ☕☕☕ (3) coffees
You doing useful work !! thanks !!
@ArtisanalANN bought ☕☕☕ (3) coffees
Enjoy.
@jsholtes bought ☕☕☕ (3) coffees
Teaching stats to civil engineer undergrads (first time teaching for me, first time for most of them too) and grasping for some good explanations of hypothesis testing, power, and CI's. Love these interactive graphics!
@notawful bought ☕☕☕ (3) coffees
Thank you for using your stats and programming gifts in such a useful, generous manner. -Jess
Mateu Servera bought ☕☕☕ (3) coffees
A job that must have cost far more coffees than we can afford you ;-). Thank you.
@cdrawn bought ☕☕☕ (3) coffees
Thank you! Such a great resource for teaching these concepts, especially CI, Power, correlation.
Julia bought ☕☕☕ (3) coffees
Fantastic work with the visualizations!
@felixthoemmes bought ☕☕☕ (3) coffees
@dalejbarr bought ☕☕☕ (3) coffees
Your work is amazing! I use your visualizations often in my teaching. Thank you.
@PsychoMouse bought ☕☕☕ (3) coffees
Excellent! Well done! SOOOO Useful!😊 🐭
Someone bought ☕☕ (2) coffees
Thanks, your work is great!!
Dan Sanes bought ☕☕ (2) coffees
this is a superb, intuitive teaching tool!
@whlevine bought ☕☕ (2) coffees
Thank you so much for these amazing visualizations. They're a great teaching tool and the allow me to show students things that it would take me weeks or months to program myself.
Someone bought ☕☕ (2) coffees
@notawful bought ☕☕ (2) coffees
Thank you for sharing your visualization skills with the rest of us! I use them frequently when teaching intro stats.
Someone bought ☕ (1) coffee
You are awesome
Thom Marchbank bought ☕ (1) coffee
Your visualisations are so useful! Thank you so much for your work.
georgina g. bought ☕ (1) coffee
thanks for helping me in my psych degree!
Someone bought ☕ (1) coffee
Thank You for this work.
Kosaku Noba bought ☕ (1) coffee
Nice visualization, I bought a cup of coffee.
Someone bought ☕ (1) coffee
Thomas bought ☕ (1) coffee
Great. Use it for teaching in psychology.
Someone bought ☕ (1) coffee
It is the best statistics visualization so far!
Ergun Pascu bought ☕ (1) coffee
AMAZING Tool!!! Thank You!
Ann Calhoun-Sauls bought ☕ (1) coffee
This has been a wonderful resource for my statistics and research methods classes. I also occassionally use it for other courses such as Theories of Personality and Social Psychology
David Britt bought ☕ (1) coffee
nicely reasoned
Mike bought ☕ (1) coffee
I appreciate your making this site available. Statistics are not in my wheelhouse, but the ability to display my data more meaningfully in my statistics class is both educational and visually appealing. Thank you!
Jayne T Jacobs bought ☕ (1) coffee
Andrew J O'Neill bought ☕ (1) coffee
Thanks for helping understand stuff!
Someone bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Shawn Hemelstrand bought ☕ (1) coffee
Thank you for this great visual. I use it all the time to demonstrate Cohen's d and why mean differences affect it's approximation.
Adele Fowler-Davis bought ☕ (1) coffee
Thank you so much for your excellent post on longitudinal models. Keep up the good work!
Stewart bought ☕ (1) coffee
This tool is awesome!
Someone bought ☕ (1) coffee
Aidan Nelson bought ☕ (1) coffee
Such an awesome page, Thank you
Someone bought ☕ (1) coffee
Ellen Kearns bought ☕ (1) coffee
Dr Nazam Hussain bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Eva bought ☕ (1) coffee
I've been learning about power analysis and effect sizes (trying to decide on effect sizes for my planned study to calculate sample size) and your Cohen's d interactive tool is incredibly useful for understanding the implications of different effect sizes!
Someone bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Thanks a lot!
Someone bought ☕ (1) coffee
Reena Murmu Nielsen bought ☕ (1) coffee
Tony Andrea bought ☕ (1) coffee
Thanks mate
Tzao bought ☕ (1) coffee
Thank you, this really helps as I am a stats idiot :)
Melanie Pflaum bought ☕ (1) coffee
Sacha Elms bought ☕ (1) coffee
Yihan Xu bought ☕ (1) coffee
Really appreciate your good work!
@stevenleung bought ☕ (1) coffee
Your visualizations really help me understand the math.
Junhan Chen bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Michael Hansen bought ☕ (1) coffee
ALEXANDER VIETHEER bought ☕ (1) coffee
mather bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Bastian Jaeger bought ☕ (1) coffee
Thanks for making the poster designs OA, I just hung two in my office and they look great!
@ValerioVillani bought ☕ (1) coffee
Thanks for your work.
Someone bought ☕ (1) coffee
Great work!
@YashvinSeetahul bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Angela bought ☕ (1) coffee
Thank you for building such excellent ways to convey difficult topics to students!
@inthelabagain bought ☕ (1) coffee
Really wonderful visuals, and such a fantastic and effective teaching tool. So many thanks!
Someone bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Yashashree Panda bought ☕ (1) coffee
I really like your work.
Ben bought ☕ (1) coffee
You're awesome. I have students in my intro stats class say, "I get it now," after using your tool. Thanks for making my job easier.
Gabriel Recchia bought ☕ (1) coffee
Incredibly useful tool!
Shiseida Sade Kelly Aponte bought ☕ (1) coffee
Thanks for the assistance for RSCH 8210.
@Benedikt_Hell bought ☕ (1) coffee
Great tools! Thank you very much!
Amalia Alvarez bought ☕ (1) coffee
@noelnguyen16 bought ☕ (1) coffee
Hi Kristoffer, many thanks for making all this great stuff available to the community!
Eran Barzilai bought ☕ (1) coffee
These visualizations are awesome! thank you for creating it
Someone bought ☕ (1) coffee
Chris SG bought ☕ (1) coffee
Very nice.
Gray Church bought ☕ (1) coffee
Thank you for the visualizations. They are fun and informative.
Qamar bought ☕ (1) coffee
Tanya McGhee bought ☕ (1) coffee
@schultemi bought ☕ (1) coffee
Neilo bought ☕ (1) coffee
Really helpful visualisations, thanks!
Someone bought ☕ (1) coffee
This is amazing stuff. Very slick.
Someone bought ☕ (1) coffee
Sarko bought ☕ (1) coffee
Thanks so much for creating this! Really helpful for being able to explain effect size to a clinician I'm doing an analysis for.
@DominikaSlus bought ☕ (1) coffee
Thank you! This page is super useful. I'll spread the word.
Someone bought ☕ (1) coffee
Melinda Rice bought ☕ (1) coffee
Thank you so much for creating these tools! As we face the challenge of teaching statistical concepts online, this is an invaluable resource.
@tmoldwin bought ☕ (1) coffee
Fantastic resource. I think you would be well served to have one page indexing all your visualizations, that would make it more accessible for sharing as a common resource.
Someone bought ☕ (1) coffee
Fantastic Visualizations! Amazing way to to demonstrate how n/power/beta/alpha/effect size are all interrelated - especially for visual learners! Thank you for creating this?
@jackferd bought ☕ (1) coffee
Incredible visualizations and the best power analysis software on R.
Cameron Proctor bought ☕ (1) coffee
Great website!
Someone bought ☕ (1) coffee
Hanah Chapman bought ☕ (1) coffee
Thank you for this work!!
Someone bought ☕ (1) coffee
Jayme bought ☕ (1) coffee
Nice explanation and visual guide of Cohen's d
Bart Comly Boyce bought ☕ (1) coffee
thank you
Dr. Mitchell Earleywine bought ☕ (1) coffee
This site is superb!
Florent bought ☕ (1) coffee
Zampeta bought ☕ (1) coffee
thank you for sharing your work.
Mila bought ☕ (1) coffee
Thank you for the website, made me smile AND smarter :O enjoy your coffee! :)
Deb bought ☕ (1) coffee
Struggling with statistics and your interactive diagram made me smile to see that someone cares enough about us strugglers to make a visual to help us out!😍
Someone bought ☕ (1) coffee
@exerpsysing bought ☕ (1) coffee
Much thanks! Visualizations are key to my learning style!
Someone bought ☕ (1) coffee
Sponsors
You can sponsor my open source work using GitHub Sponsors and have your name shown here.
Backers ✨❤️
Questions & Comments
Please use GitHub Discussions for any questions related to this post, or open an issue on GitHub if you've found a bug or wan't to make a feature request.
Archived Comments (38)
Hi,
Great script but I can't run it
It appears this error. Can you help me? Thkx a lot
df <- PubMedTrend(query)
Searching for: medicine[tw]
| | 0%Space required after the Public Identifier
SystemLiteral " or ' expected
SYSTEM or PUBLIC, the URI is missing
Error: 1: Space required after the Public Identifier
2: SystemLiteral " or ' expected
3: SYSTEM or PUBLIC, the URI is missing
Having the same problem...was solved by changing the URL in the retrieval loop to include https instead of http.
Hello Kristoffer i am trying to use your code to extract the # of citation from a lot of articles in PubMed but i get this error
""Error in function (type, msg, asError = TRUE) :
SSL certificate problem: unable to get local issuer certificate""
Maybe you could help?i dont have any idea what this error is,although i search it on the WEB
thnx in advance
Thank you, this script is great!
I had one problem with it however: Line 62, where you load total_table.csv
You wrote
tmp <- getURL("https://raw.github.com/rpsy...")
but for me this only returned an emtpy character ""
What worked is changing the URL to
tmp <- getURL("https://raw.githubuserconte...")
If I missed something: sorry! If not, it might reflect an internal change at github and you might want to update the file.
All the best,
Florian
Hi Kristoffer,
very nice script, however it does not seem to work for me. I tried several queries but they never work. Always the same error, see below. The debug does not point to a specific line so I have no clue where to start. Maybe you have suggestion? All packages are up to date.
> source("PubMedTrend.R")
Loading required package: RCurl
Loading required package: bitops
Loading required package: XML
Loading required package: plyr
> query <- c("biology" = "biology[tw]")
> df <- PubMedTrend(query)
Searching for: biology[tw]
|=======================================================================================================| 100%
Error in read.table(file = file, header = header, sep = sep, quote = quote, :
no lines available in input
Called from: read.csv(text = tmp)
Browse[1]>
Morning Kristoffer, I need a Pubmed count done on publications on QEEG and ERP in the past 10 years with a graph. I have one, but do not know where the author got it or how to reference as it is from the user group Facebook page and not an academic resource. Could you pls assist. Perhaps if you draw up a new similar statistic for me you could assist in how to reference it correctly. Hope I am understanding correctly what you do and that you can assist in this for me. Pls let me know if I needed to compensate you. Your work is really interesting and of huge use.
You may want to contribute your code to one of these two R packages:
https://github.com/ropensci...
https://github.com/ropensci...
Hi,
thanks a lot for this function.
I tried to run it, but encountered this error
df <- PubMedTrend(query, 1990,2005)
Searching for: medicine[tw]
|======================================================================| 100%
Searching for: psychology[tw]
|======================================================================| 100%
Searching for: biology[tw]
|======================================================================| 100%
Error in match(df$year, total.table$year) :
object 'total.table' not found
Do you have an idea why that is?
Thanks a lot,
Best,
JR
The script didn't find the total.table file. I've updated the script to avoid this problem. Now it'll download the total.table data-file from my GitHub repo.
Hi, great code. I am a bit confused though. You are calculating the number of publications per million by calculating the relative counts * 10,000. Is this not the number of publication per 10,000 or am I not getting the point?
You are absolutely right. Thanks for notifying me. Must have been tired when wrote that :). I've changed the function so it'll return the relative counts as percentages. I figured it's better to do any computations outside the main procedure, if it's needed.
Great script, thanks for writing it! One caveat I noticed when I was running/modifying it. I was searching for a count of "Dangeardiella macrospora". The API call (no years) shows that there are 109 hits:
http://eutils.ncbi.nlm.nih....
*but*, searching at PubMed shows that the search result should actually be zero (since "Dangeardiella" is not found in PubMed, they conveniently just discard that search term):
http://www.ncbi.nlm.nih.gov...
To ask that PubMed not discard terms, you need to append "[All fields]". For example:
http://www.ncbi.nlm.nih.gov...
http://eutils.ncbi.nlm.nih....[All%20fields]
Hope that's helpful!
The last link in my comment is slightly broken (square brackets got lost). The correct link is here: http://tinyurl.com/km238fq
Thanks for posting this Andrew! I've opened an issue regarding this on my github repo. I will look into it when have some time over.
Nice one Kris,
I am puzzled as to why the total counts for 2011 onwards to 2013 are NAs I cannot find any fault in the code and my system date is on the correct year! so for example if I query for 2009 to 2012 I get:
.idyearcounttotal_countrelative
1App20091507786831.9263295
2App20101696800982.4849360
3App2011 166NA NA
4App2012194NA NA
5Rev2009147786830.1797907
6Rev2010216800980.3087790
7Rev2011 31NA NA
8Rev201223NA NA
Hi Andrews, the script didn't include total values for 2011 onwards. The totals were fetched from this table http://www.nlm.nih.gov/bsd/.... It's been updated now, and so has my script.
Great piece of code. I am trying to extend the publication year range from1970 to 2012 but the total_table only goes from 1947 to 2009 (as commented in your code), correct?
Is there a way I can download or update the table to include 2012?
Hi Fulton. Here's two different ways to update the total_table https://github.com/rpsychol...
Amazing. I'm wondering how it might be applied to ProMED-mail archives now.....
In the meantime, however...a much easier problem. How would one plot the data with a log2 y-axis? I'm finding the learning curve for gglot2 is much steeper than that of R itself....
Hi Robert. Did you figure out how to plot the data with a log2 y-axis? If not, you can use something like this: scale_y_continuous(trans=log2_trans()), this is using the log_trans from the "scales"-packages, so you need that package loaded.
Very nice, any plans to post it as a package on CRAN?
Hi Ricardo, maybe In the future, but it's not really something I've thought about!
Whilst I actually like this publish, I believe there was an spelling error near towards the finish of the third section.
Thanks for notifying me, it's been sorted!
I get this error after it has finished searching when trying to run your script (using rstudio):
Error in readChar(con, 5L, useBytes = TRUE) : cannot open the connection
In addition: Warning message:
In readChar(con, 5L, useBytes = TRUE) :
cannot open compressed file 'total_table', probable reason 'No such file or directory'
Any idea what is going on?
I get a similar area in that it can't find 'total.table', have redownloaded, tried to rename total.table to total_table...
Using Mac OS X 10.8.4
Kris,
thanks for sharing this script. It's exactly what I was trying to do, with a lot of pain.
However I don't find any file other than PubMedTrend.r and I get the same error as Wouter. Also, I don't find RunPubMed.R mentioned by Michael MacAskill.
I assume both come from the same zip file, but where is the link ?
Cheers
D
Hi, Dani. I have moved everything to moved the script over to GitHub so you should find everything here.
Works brilliantly, in a different league performance and reliability-wise compared to RISmed.
One possible error in line 28 of RunPubMed.R
Should:
df.hits <- PubMedTrend()
actually be:
df.hits <- PubTotalHits()
Cheers.
Glad you liked it! You are correct about the error on line 28, thanks for spotting it!
Hi Kris,
Thank you for this fine and incredibly useful script.
Unfortunately it does not work with the recent version R (2.15) since some interfaces seem to have changed. When running the example script I receive an error message which seems to distresst already some R users:
> source("PubMedTrend.R")
Error in source("PubMedTrend.R") :
7 arguments passed to .Internal(identical) which requires 6
http://r.789695.n4.nabble.c...
Since I am not too experienced yet, I do not have an idea which of the function calls .Internal() . If you have a guess and give a hint I would try bugfixing.
Best regards,
Robert
Hi Robert,
Unfortunately I couldn't replicate you error on R 2.15.0. Are you using the latest versions of "plyr", "XML" and "Rcurl"?.
I'm not explicitly calling .Internal in my script, but you could try running traceback() to get some more information on the error.
Hi Kris,
Strangely, it seems that neither your script nor R itself is the troublemaker but RStudio - sorry for not mentioning the use of an alternative editor first. Due to the above given link I thought that the error ist related to the updated version of R.
When directly inputting your script into RStudio (by loading -> Strg A -> Run) everything works well and the given examples are executeable.
When loading your script with source followed by traceback():
> source("PubMedTrend.R")
Error in source("PubMedTrend.R") :
7 arguments passed to .Internal(identical) which requires 6
> traceback()
2: source("PubMedTrend.R")
1: source("PubMedTrend.R")
Nevertheless, using the native R console no error is thrown even when loading the script with source().
Sorry for the confusion and thank you again for providing the code!
Thanks for posting your solution, I'm glad you got it to work. It's strange though, it runs just fine in RStudio for me.
Thanks for sharing Kris, particularly since the code even includes thoughtful validation checking!
Looking forward to trying this out, but will be a few days I think. An advantage of the RISmed approach was that (when it worked...) entire records were returned. This meant that further analyses could be done, above just the simple raw counts (e.g. examining trends within a particular journal). I'll be happy to at least get reliable count data, but does this method lend itself to getting individual record-level information as well?
Cheers,
Michael
It's possible to extend this method to download complete records in XML. This script actually started out with that functionality, but it seemed a bit unnecessary to download the complete records when I mostly wanted to look at yearly counts.
I will release a version of my script that will download complete records, I only need to add a function to batch download articles, since PubMed has got a retrieval cap of 10k articles.
Thanks for commenting,
Kris
