Why linear mixed-effects models are probably not the solution to your missing data problems
Linear mixed-effects models are often used for their ability to handle missing data using maximum likelihood estimation. In this post I will present a simple example of when the LMM fails, and illustrate two MNAR sensitivity analyses: the pattern-mixture method and the joint model (shared parameter model). This post is based on a small example from my PhD thesis.
MCAR, MAR, and MNAR missing data
D. B. Rubin (1976) presented three types of missing data mechanisms: missing completely at random (MCAR), missing at random (MAR), missing not at random (MNAR). LMMs provide unbiased estimates under MAR missingness. If we have the complete outcome variable (which is made up of the observed data and the missing values ) and a missing data indicator (D. B. Rubin 1976; R. J. Little and Rubin 2014; Schafer and Graham 2002), then we can write the MCAR and MAR mechanisms as,
If the missingness depends on , the missing values in , then the mechanism is MNAR. MCAR and MAR are called ignorable because the precise model describing the missing data process is not needed. In theory, valid inference under MNAR missingness requires specifying a joint distribution for both the data and the missingness mechanisms (R. J. A. Little 1995). There are no ways to test if the missing data are MAR or MNAR (Molenberghs et al. 2008; Rhoads 2012), and it is therefore recommended to perform sensitivity analyses using different MNAR mechanisms (Schafer and Graham 2002; R. J. A. Little 1995; Hedeker and Gibbons 1997).
LMMs and missing data
LMMs are frequently used by researchers to try to deal with missing data problems. However, researchers frequently misunderstand the MAR assumption and often fail to build a model that would make the assumption more plausible. Sometimes you even see researchers using tests, e.g., Little’s MCAR test, to prove that the missing data mechanisms is either MCAR or MAR and hence ignorable—which is clearly a misunderstanding and builds on faulty logic.
A common problem is that researchers do not include covariates that potentially predict dropout. Thus, it is assumed that missingness only depend on the previously observed values of the outcome. This is quite a strong assumption. A related misunderstanding, is that the LMM’s missing data assumption is more liberal as it allows for participants’ slopes to vary. It is sometimes assumed tat if a random slope is included in the model it can also be used to satisfy the MAR assumption. Clearly, it would be very practical if the inclusion of random slopes would allow missingness to depend on patients’ latent change over time. Because it is probably true that some participants’ dropout is related to their symptom’s rate of change over time. Unfortunately, the random effects are latent variables and not observed variables—hence, such a missingness mechanism would also be MNAR (R. J. A. Little 1995). The figure below illustrates the MAR, outcome-based MNAR, and random coefficient-based MNAR mechanisms.
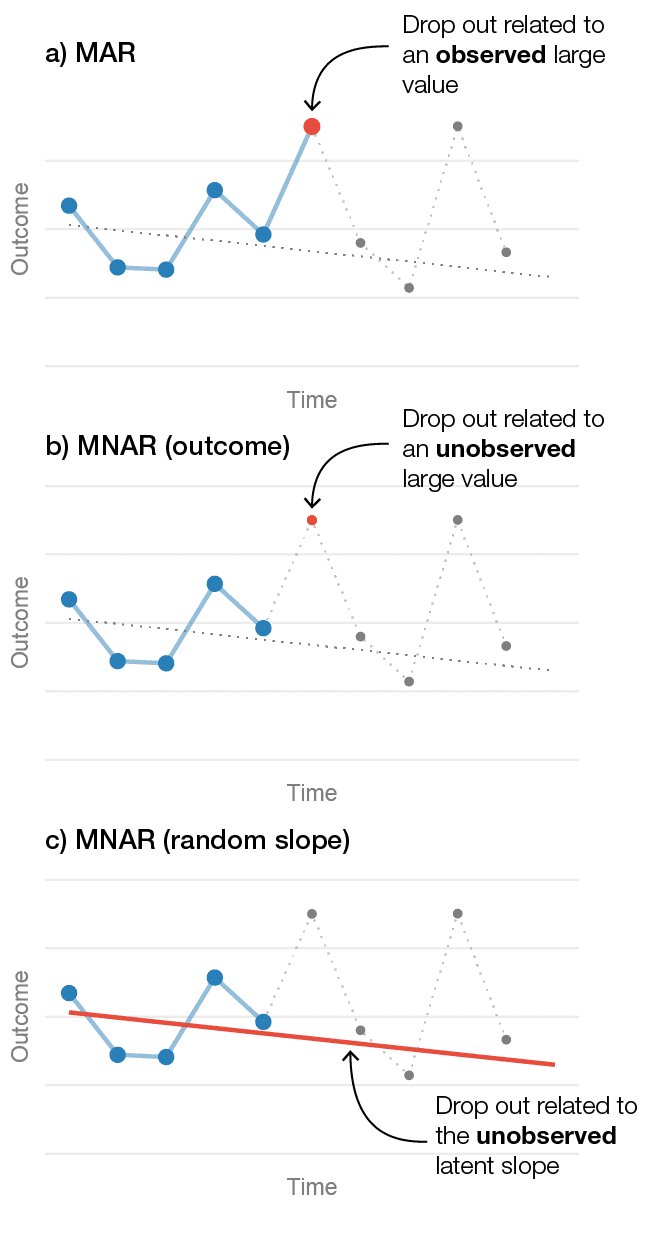
Figure 1. Three different drop out mechanisms in longitudinal data from one patient. a) Illustrates a MAR mechanism where the patient's likelihood of dropping out is related to an observed large value. b) Shows an outcome-related MNAR mechanism, where dropout is related to a large unobserved value. c) Shows a random-slope MNAR mechanism where the likelihood of dropping out is related to the patient's unobserved slope.
Let’s generate some data
To illustrate these concepts let’s generate data from a two-level LMM with random intercept and slopes, and included a MNAR missing data mechanism where the likelihood of dropping out depended on the patient-specific random slopes. Moreover, let’s assume that the missingness differs between the treatment and control group. This isn’t that unlikely in unblinded studies (e.g., wait-list controls).
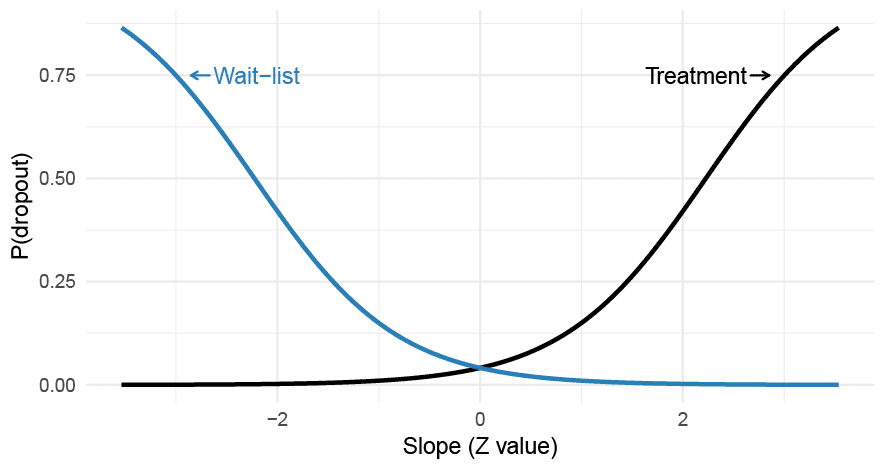
Figure 2. A differential MNAR dropout process where the probability of dropping out from a trial depends on the patient-specific slopes which interact with the treatment allocation. The probability of dropout is assumed to be constant over time.
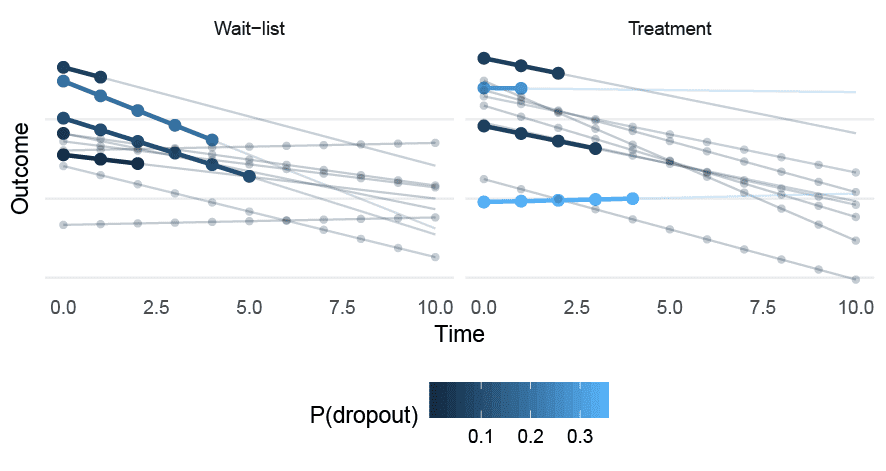
Figure 3. A sample of patients drawn from the MNAR (random slope) data-generating process. Circles represent complete observations; the bold line represents the slope before dropping out. P(dropout) gives the probability of dropout, which is assumed to be constant at all time points.
The equations for the dropout can be written as,
The R code is quite simple,
Now let’s draw a large sample from this model (1000 participants per group), and fit a typical longitudinal LMM using both the complete outcome variable and the incomplete (MNAR) outcome variable.
Here are the results (click on “SHOW” to see the output).
We can see that the slope difference is -0.25 for the complete data and much larger for the LMM with missing data (-1.14).
A Pattern-mixture model
A simple extension of the classical LMM is a pattern-mixture model. This is a simple model where we allow the slope to differ within subgroups of different dropout patterns. The simplest pattern is to group the participants into two subgroups dropouts (1) or completers (0), and include this dummy variable in the model.
As you can see in the output, we now have a bunch of new coefficients. In order to get the marginal treatment effect we need to average over the dropout patterns. There are several ways to do this, we could just calculate a weighted average manually. For example, the outcome at posttest in the control group is
To estimate the treatment effect we’d need to repeat this for the treatment group and take the difference. However, we’d also need to calculate the standard errors (e.g., using the delta method). An easier option is to just specify the linear contrast we are interest in.
This tells us that the difference between the groups at posttest is estimated to be -4.65. This is considerably smaller than the estimate from the classical LMM, but still larger then for the complete data. We could accomplish to same thing using emmeans package.
Fitting a joint model
The pattern-mixture model was an improvement, but it didn’t completely recover the treatment effect under the random slope MNAR model. We can actually fit a model that allows dropout to be related to the participants’ random slopes. To accomplish this we combine a survival model for the dropout process and an LMM for the longitudinal outcome.
We can see from the output that the estimate of the treatment effect is really close to the estimate from the complete data (-0.23 vs -0.25). There’s only one small problem with the joint model and that is that we almost never know what the correct model is…
A small simulation
Now let’s run a small simulation to show the consequences of this random-slope dependent MNAR scenario. We’ll do a study with 11 time points, 150 participants per group, a variance ratio of 0.02, and pretest ICC = 0.6, with a correlation between intercept and slopes of -0.5. There will be a “small” effect in favor of the treatment of . The following models will be compared:
- LMM (MAR): a classical LMM assuming that the dropout was MAR.
- GEE: a generalized estimating equation model.
- LMM (PM): an LMM using a pattern-mixture approach. Two patterns were used; either “dropout” or “completer”, and the results were averaged over the two patterns.
- JM: A joint model that correctly allowed the dropout to be related to the random slopes.
- LMM with complete data: an LMM fit to the complete data without any missingness.
I will not post all code here; the complete code for this post can be found on GitHub. Here’s a snippet showing the code that was used to fit the models.
Results
The table and figure below shows how much the treatment effects differ. We can see that LMMs are badly biased under this missing data scenario; the treatment effect is much larger than it should be (Cohen’s d: -0.7 vs. -0.2). The pattern-mixture approach improves the situation, and the joint model recovers the true effect. Since the sample size is large, the bias under the MAR assumption leads to the LMM’s CIs having extremely bad coverage. Moreover, under the assumption of no treatment effect the MAR LMM’s type I errors are very high (83%), whereas the pattern-mixture and joint model are closer to the nominal levels.
| Model | M(Est.) | Rel. bias | d | Power | CI coverage | Type I error |
|---|---|---|---|---|---|---|
| MAR | -11.84 | 274.38 | -0.74 | 1.00 | 0.02 | 0.83 |
| PM | -5.39 | 70.47 | -0.34 | 0.64 | 0.84 | 0.10 |
| GEE | -11.19 | 253.98 | -0.70 | 1.00 | 0.06 | 0.71 |
| JM | -3.18 | 0.59 | -0.20 | 0.28 | 0.93 | 0.07 |
| Complete | -3.21 | 1.44 | -0.20 | 0.38 | 0.95 | 0.05 |
Note: MAR = missing at random; LMM = linear mixed-effects model; GEE = generalized estimating equation; JM = joint model; PM = pattern mixture; Est. = mean of the estimated effects; Rel. bias = relative bias of Est.; d = mean of the Cohen’s d estimates.
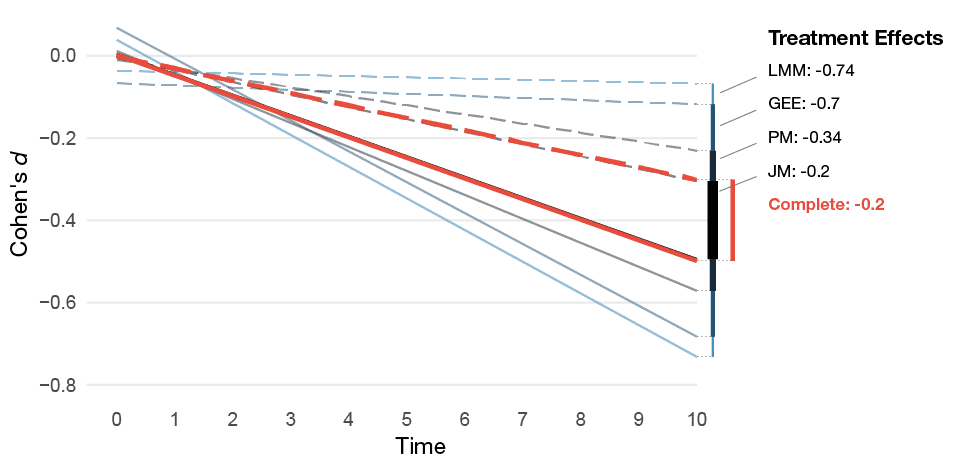
Figure 3. Mean of the estimated treatment effect from the MNAR missing data simulations for the different models. The dashed lines represents the control group's estimated average slope and the solid lines the treatment group's average slope.
Summary
This example is purposely quite extreme. However, even if the MNAR mechanism would be weaker, the LMM will yield biased estimates of the treatment effect. The assumption that dropout might be related to patients’ unobserved slopes is not unreasonable. However, fitting a joint model is often not feasible as we do not know the true missingness mechanism. I included it just to illustrate what is required to avoid bias under a plausible MNAR mechanism. In reality, the patients’ likelihood of dropping out is likely an inseparable mix of various degrees of MCAR, MAR, and MNAR mechanisms. The only sure way of avoiding bias would be to try to acquire data from all participants—and when that fails, perform sensitivity analyses using reasonable assumptions of the missingness mechanisms.
References
Hedeker, Donald, and Robert D Gibbons. 1997. “Application of Random-Effects Pattern-Mixture Models for Missing Data in Longitudinal Studies.” Psychological Methods 2 (1): 64–78. doi:10.1037/1082-989X.2.1.64.
Little, Roderick J. A. 1995. “Modeling the Drop-Out Mechanism in Repeated-Measures Studies.” Journal of the American Statistical Association 90 (431): 1112–21. doi:10.1080/01621459.1995.10476615.
Little, Roderick JA, and Donald B Rubin. 2014. Statistical Analysis with Missing Data. Vol. 333. John Wiley & Sons.
Molenberghs, Geert, Caroline Beunckens, Cristina Sotto, and Michael G. Kenward. 2008. “Every Missingness Not at Random Model Has a Missingness at Random Counterpart with Equal Fit.” Journal of the Royal Statistical Society: Series B (Statistical Methodology) 70 (2): 371–88. doi:10.1111/j.1467-9868.2007.00640.x.
Rhoads, Christopher H. 2012. “Problems with Tests of the Missingness Mechanism in Quantitative Policy Studies.” Statistics, Politics, and Policy 3 (1). doi:10.1515/2151-7509.1012.
Rubin, Donald B. 1976. “Inference and Missing Data.” Biometrika 63 (3): 581–92. doi:10.1093/biomet/63.3.581.
Schafer, Joseph L., and John W. Graham. 2002. “Missing Data: Our View of the State of the Art.” Psychological Methods 7 (2): 147–77. doi:10.1037//1082-989X.7.2.147.
Written by Kristoffer Magnusson, a researcher in clinical psychology. You should follow him on Twitter and come hang out on the open science discord Git Gud Science.
Published July 09, 2020 (View on GitHub)
Buy Me A Coffee
A huge thanks to the 152 supporters who've bought me a 361 coffees!
Jason Rinaldo bought ☕☕☕☕☕☕☕☕☕☕ (10) coffees
I've been looking for applets that show this for YEARS, for demonstrations for classes. Thank you so much! Students do not need to tolarate my whiteboard scrawl now. I'm sure they'd appreciate you, too.l
JDMM bought ☕☕☕☕☕ (5) coffees
You finally helped me understand correlation! Many, many thanks... 😄
@VicCazares bought ☕☕☕☕☕ (5) coffees
Good stuff! It's been so helpful for teaching a Psych Stats class. Cheers!
Dustin M. Burt bought ☕☕☕☕☕ (5) coffees
Excellent and informative visualizations!
Someone bought ☕☕☕☕☕ (5) coffees
@metzpsych bought ☕☕☕☕☕ (5) coffees
Always the clearest, loveliest simulations for complex concepts. Amazing resource for teaching intro stats!
Ryo bought ☕☕☕☕☕ (5) coffees
For a couple years now I've been wanting to create visualizations like these as a way to commit these foundational concepts to memory. But after finding your website I'm both relieved that I don't have to do that now and pissed off that I couldn't create anything half as beautiful and informative as you have done here. Wonderful job.
Diarmuid Harvey bought ☕☕☕☕☕ (5) coffees
You have an extremely useful site with very accessible content that I have been using to introduce colleagues and students to some of the core concepts of statistics. Keep up the good work, and thanks!
Michael Hansen bought ☕☕☕☕☕ (5) coffees
Keep up the good work!
Michael Villanueva bought ☕☕☕☕☕ (5) coffees
I wish I could learn more from you about stats and math -- you use language in places that I do not understand. Cohen's D visualizations opened my understanding. Thank you
Someone bought ☕☕☕☕☕ (5) coffees
Thank you, Kristoffer
Pål from Norway bought ☕☕☕☕☕ (5) coffees
Great webpage, I use it to illustrate several issues when I have a lecture in research methods. Thanks, it is really helpful for the students:)
@MAgrochao bought ☕☕☕☕☕ (5) coffees
Joseph Bulbulia bought ☕☕☕☕☕ (5) coffees
Hard to overstate the importance of this work Kristoffer. Grateful for all you are doing.
@TDmyersMT bought ☕☕☕☕☕ (5) coffees
Some really useful simulations, great teaching resources.
@lakens bought ☕☕☕☕☕ (5) coffees
Thanks for fixing the bug yesterday!
@LinneaGandhi bought ☕☕☕☕☕ (5) coffees
This is awesome! Thank you for creating these. Definitely using for my students, and me! :-)
@ICH8412 bought ☕☕☕☕☕ (5) coffees
very useful for my students I guess
@KelvinEJones bought ☕☕☕☕☕ (5) coffees
Preparing my Master's student for final oral exam and stumbled on your site. We are discussing in lab meeting today. Coffee for everyone.
Someone bought ☕☕☕☕☕ (5) coffees
What a great site
@Daniel_Brad4d bought ☕☕☕☕☕ (5) coffees
Wonderful work!
David Loschelder bought ☕☕☕☕☕ (5) coffees
Terrific work. So very helpful. Thank you very much.
@neilmeigh bought ☕☕☕☕☕ (5) coffees
I am so grateful for your page and can't thank you enough!
@giladfeldman bought ☕☕☕☕☕ (5) coffees
Wonderful work, I use it every semester and it really helps the students (and me) understand things better. Keep going strong.
Dean Norris bought ☕☕☕☕☕ (5) coffees
Sal bought ☕☕☕☕☕ (5) coffees
Really super useful, especially for teaching. Thanks for this!
dde@paxis.org bought ☕☕☕☕☕ (5) coffees
Very helpful to helping teach teachers about the effects of the Good Behavior Game
@akreutzer82 bought ☕☕☕☕☕ (5) coffees
Amazing visualizations! Thank you!
@rdh_CLE bought ☕☕☕☕☕ (5) coffees
So good!
Amanda Sharples bought ☕☕☕ (3) coffees
Soyol bought ☕☕☕ (3) coffees
Someone bought ☕☕☕ (3) coffees
Kenneth Nilsson bought ☕☕☕ (3) coffees
Keep up the splendid work!
@jeremywilmer bought ☕☕☕ (3) coffees
Love this website; use it all the time in my teaching and research.
Someone bought ☕☕☕ (3) coffees
Powerlmm was really helpful, and I appreciate your time in putting such an amazing resource together!
DR AMANDA C DE C WILLIAMS bought ☕☕☕ (3) coffees
This is very helpful, for my work and for teaching and supervising
Georgios Halkias bought ☕☕☕ (3) coffees
Regina bought ☕☕☕ (3) coffees
Love your visualizations!
Susan Evans bought ☕☕☕ (3) coffees
Thanks. I really love the simplicity of your sliders. Thanks!!
@MichaMarie8 bought ☕☕☕ (3) coffees
Thanks for making this Interpreting Correlations: Interactive Visualizations site - it's definitely a great help for this psych student! 😃
Zakaria Giunashvili, from Georgia bought ☕☕☕ (3) coffees
brilliant simulations that can be effectively used in training
Someone bought ☕☕☕ (3) coffees
@PhysioSven bought ☕☕☕ (3) coffees
Amazing illustrations, there is not enough coffee in the world for enthusiasts like you! Thanks!
Cheryl@CurtinUniAus bought ☕☕☕ (3) coffees
🌟What a great contribution - thanks Kristoffer!
vanessa moran bought ☕☕☕ (3) coffees
Wow - your website is fantastic, thank you for making it.
Someone bought ☕☕☕ (3) coffees
mikhail.saltychev@gmail.com bought ☕☕☕ (3) coffees
Thank you Kristoffer This is a nice site, which I have been used for a while. Best Prof. Mikhail Saltychev (Turku University, Finland)
Someone bought ☕☕☕ (3) coffees
Ruslan Klymentiev bought ☕☕☕ (3) coffees
@lkizbok bought ☕☕☕ (3) coffees
Keep up the nice work, thank you!
@TELLlab bought ☕☕☕ (3) coffees
Thanks - this will help me to teach tomorrow!
SCCT/Psychology bought ☕☕☕ (3) coffees
Keep the visualizations coming!
@elena_bolt bought ☕☕☕ (3) coffees
Thank you so much for your work, Kristoffer. I use your visualizations to explain concepts to my tutoring students and they are a huge help.
A random user bought ☕☕☕ (3) coffees
Thank you for making such useful and pretty tools. It not only helped me understand more about power, effect size, etc, but also made my quanti-method class more engaging and interesting. Thank you and wish you a great 2021!
@hertzpodcast bought ☕☕☕ (3) coffees
We've mentioned your work a few times on our podcast and we recently sent a poster to a listener as prize so we wanted to buy you a few coffees. Thanks for the great work that you do!Dan Quintana and James Heathers - Co-hosts of Everything Hertz
Cameron Proctor bought ☕☕☕ (3) coffees
Used your vizualization in class today. Thanks!
eshulman@brocku.ca bought ☕☕☕ (3) coffees
My students love these visualizations and so do I! Thanks for helping me make stats more intuitive.
Someone bought ☕☕☕ (3) coffees
Adrian Helgå Vestøl bought ☕☕☕ (3) coffees
@misteryosupjoo bought ☕☕☕ (3) coffees
For a high school teacher of psychology, I would be lost without your visualizations. The ability to interact and manipulate allows students to get it in a very sticky manner. Thank you!!!
Chi bought ☕☕☕ (3) coffees
You Cohen's d post really helped me explaining the interpretation to people who don't know stats! Thank you!
Someone bought ☕☕☕ (3) coffees
You doing useful work !! thanks !!
@ArtisanalANN bought ☕☕☕ (3) coffees
Enjoy.
@jsholtes bought ☕☕☕ (3) coffees
Teaching stats to civil engineer undergrads (first time teaching for me, first time for most of them too) and grasping for some good explanations of hypothesis testing, power, and CI's. Love these interactive graphics!
@notawful bought ☕☕☕ (3) coffees
Thank you for using your stats and programming gifts in such a useful, generous manner. -Jess
Mateu Servera bought ☕☕☕ (3) coffees
A job that must have cost far more coffees than we can afford you ;-). Thank you.
@cdrawn bought ☕☕☕ (3) coffees
Thank you! Such a great resource for teaching these concepts, especially CI, Power, correlation.
Julia bought ☕☕☕ (3) coffees
Fantastic work with the visualizations!
@felixthoemmes bought ☕☕☕ (3) coffees
@dalejbarr bought ☕☕☕ (3) coffees
Your work is amazing! I use your visualizations often in my teaching. Thank you.
@PsychoMouse bought ☕☕☕ (3) coffees
Excellent! Well done! SOOOO Useful!😊 🐭
Dan Sanes bought ☕☕ (2) coffees
this is a superb, intuitive teaching tool!
@whlevine bought ☕☕ (2) coffees
Thank you so much for these amazing visualizations. They're a great teaching tool and the allow me to show students things that it would take me weeks or months to program myself.
Someone bought ☕☕ (2) coffees
@notawful bought ☕☕ (2) coffees
Thank you for sharing your visualization skills with the rest of us! I use them frequently when teaching intro stats.
Andrew J O'Neill bought ☕ (1) coffee
Thanks for helping understand stuff!
Someone bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Shawn Hemelstrand bought ☕ (1) coffee
Thank you for this great visual. I use it all the time to demonstrate Cohen's d and why mean differences affect it's approximation.
Adele Fowler-Davis bought ☕ (1) coffee
Thank you so much for your excellent post on longitudinal models. Keep up the good work!
Stewart bought ☕ (1) coffee
This tool is awesome!
Someone bought ☕ (1) coffee
Aidan Nelson bought ☕ (1) coffee
Such an awesome page, Thank you
Someone bought ☕ (1) coffee
Ellen Kearns bought ☕ (1) coffee
Dr Nazam Hussain bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Eva bought ☕ (1) coffee
I've been learning about power analysis and effect sizes (trying to decide on effect sizes for my planned study to calculate sample size) and your Cohen's d interactive tool is incredibly useful for understanding the implications of different effect sizes!
Someone bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Thanks a lot!
Someone bought ☕ (1) coffee
Reena Murmu Nielsen bought ☕ (1) coffee
Tony Andrea bought ☕ (1) coffee
Thanks mate
Tzao bought ☕ (1) coffee
Thank you, this really helps as I am a stats idiot :)
Melanie Pflaum bought ☕ (1) coffee
Sacha Elms bought ☕ (1) coffee
Yihan Xu bought ☕ (1) coffee
Really appreciate your good work!
@stevenleung bought ☕ (1) coffee
Your visualizations really help me understand the math.
Junhan Chen bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Michael Hansen bought ☕ (1) coffee
ALEXANDER VIETHEER bought ☕ (1) coffee
mather bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Bastian Jaeger bought ☕ (1) coffee
Thanks for making the poster designs OA, I just hung two in my office and they look great!
@ValerioVillani bought ☕ (1) coffee
Thanks for your work.
Someone bought ☕ (1) coffee
Great work!
@YashvinSeetahul bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Angela bought ☕ (1) coffee
Thank you for building such excellent ways to convey difficult topics to students!
@inthelabagain bought ☕ (1) coffee
Really wonderful visuals, and such a fantastic and effective teaching tool. So many thanks!
Someone bought ☕ (1) coffee
Someone bought ☕ (1) coffee
Yashashree Panda bought ☕ (1) coffee
I really like your work.
Ben bought ☕ (1) coffee
You're awesome. I have students in my intro stats class say, "I get it now," after using your tool. Thanks for making my job easier.
Gabriel Recchia bought ☕ (1) coffee
Incredibly useful tool!
Shiseida Sade Kelly Aponte bought ☕ (1) coffee
Thanks for the assistance for RSCH 8210.
@Benedikt_Hell bought ☕ (1) coffee
Great tools! Thank you very much!
Amalia Alvarez bought ☕ (1) coffee
@noelnguyen16 bought ☕ (1) coffee
Hi Kristoffer, many thanks for making all this great stuff available to the community!
Eran Barzilai bought ☕ (1) coffee
These visualizations are awesome! thank you for creating it
Someone bought ☕ (1) coffee
Chris SG bought ☕ (1) coffee
Very nice.
Gray Church bought ☕ (1) coffee
Thank you for the visualizations. They are fun and informative.
Qamar bought ☕ (1) coffee
Tanya McGhee bought ☕ (1) coffee
@schultemi bought ☕ (1) coffee
Neilo bought ☕ (1) coffee
Really helpful visualisations, thanks!
Someone bought ☕ (1) coffee
This is amazing stuff. Very slick.
Someone bought ☕ (1) coffee
Sarko bought ☕ (1) coffee
Thanks so much for creating this! Really helpful for being able to explain effect size to a clinician I'm doing an analysis for.
@DominikaSlus bought ☕ (1) coffee
Thank you! This page is super useful. I'll spread the word.
Someone bought ☕ (1) coffee
Melinda Rice bought ☕ (1) coffee
Thank you so much for creating these tools! As we face the challenge of teaching statistical concepts online, this is an invaluable resource.
@tmoldwin bought ☕ (1) coffee
Fantastic resource. I think you would be well served to have one page indexing all your visualizations, that would make it more accessible for sharing as a common resource.
Someone bought ☕ (1) coffee
Fantastic Visualizations! Amazing way to to demonstrate how n/power/beta/alpha/effect size are all interrelated - especially for visual learners! Thank you for creating this?
@jackferd bought ☕ (1) coffee
Incredible visualizations and the best power analysis software on R.
Cameron Proctor bought ☕ (1) coffee
Great website!
Someone bought ☕ (1) coffee
Hanah Chapman bought ☕ (1) coffee
Thank you for this work!!
Someone bought ☕ (1) coffee
Jayme bought ☕ (1) coffee
Nice explanation and visual guide of Cohen's d
Bart Comly Boyce bought ☕ (1) coffee
thank you
Dr. Mitchell Earleywine bought ☕ (1) coffee
This site is superb!
Florent bought ☕ (1) coffee
Zampeta bought ☕ (1) coffee
thank you for sharing your work.
Mila bought ☕ (1) coffee
Thank you for the website, made me smile AND smarter :O enjoy your coffee! :)
Deb bought ☕ (1) coffee
Struggling with statistics and your interactive diagram made me smile to see that someone cares enough about us strugglers to make a visual to help us out!😍
Someone bought ☕ (1) coffee
@exerpsysing bought ☕ (1) coffee
Much thanks! Visualizations are key to my learning style!
Someone bought ☕ (1) coffee
Sponsors
You can sponsor my open source work using GitHub Sponsors and have your name shown here.
Backers ✨❤️
Questions & Comments
Please use GitHub Discussions for any questions related to this post, or open an issue on GitHub if you've found a bug or wan't to make a feature request.
Webmentions
0 0
(Webmentions sent before 2021 will unfortunately not show up here.)